Serviços Personalizados
Artigo
Links relacionados
Compartilhar
Brazilian Journal of Oral Sciences
versão On-line ISSN 1677-3225
Braz. J. Oral Sci. vol.13 no.3 Piracicaba Jul./Set. 2014
ORIGINAL ARTICLE
Using molecular markers to assess Streptococcus mutans variability and the biological risk for caries
Ivana Froede NeivaI; Mônica MoreiraI; Renata Rodrigues GomesII; Debora KlisiowiczII; Ricardo Lehtonen Rodrigues SouzaIII; Vânia Aparecida VicenteI,II
I Universidade Federal do Paraná – UFPR, Department of Chemical Engineering, Areas of Engineer of Bioprocess and Biotechnology Curitiba, PR, Brazil
II Universidade Federal do Paraná – UFPR, Department of Basic Pathology, Areas of Microbiology, Parasitology and Pathology, Biological Sciences, Curitiba, PR, Brazil
III Universidade Federal do Paraná – UFPR, Department of Genetics, Curitiba, PR, Brazil
ABSTRACT
Aim: To characterize the genetic variability of Streptococcus mutans isolates and to correlate this variability with different colonization profiles observed during dental caries in a sample of children. Methods: S. mutans samples were isolated from the saliva of 30 children with varying histories of dental caries, and they were characterized according to morphological and biochemical markers and the sequences of their 16S-23S intergenic spacer region. The genetic variability of the isolates was first assessed using Random Amplified Polymorphic DNA (RAPD) markers. Next, the isolates were differentiated by sequencing a specific region of the gene encoding the enzyme glucosyltransferase B (gtfB). Results: Characterization using RAPD markers uncovered significant genetic variability among the samples and indicated the existence of clusters, which allowed us to reconstruct both the origin and clinical history of the disease. By sequencing the 16S-23S intergenic region, it was found that all of the isolates belonged to the species S. mutans. Based on the genetic similarity of the isolates and pattern of amino acid variations identified by partial sequencing of the gtfB gene, base-pair changes were identified and correlated with different virulence patterns among the isolates. Conclusions: The partial sequencing of the gtfB gene can be a useful tool for elucidating the colonization patterns of S. mutans. As amino acid variations are likely to be correlated with differences in biological risk, molecular characterization, such as that described in this paper, could be the key for assessing the development of dental caries in children.
Keywords: Streptococcus mutans; dental caries; Random amplified polymorphic DNA technique; glucosyltransferase.
Introduction
Caries is an infectious and transmissible disease associated with bacterial colonization of dental surfaces1. Due to its multifactorial nature and microbial origin, the severity and prevalence of caries can be greatly affected by the endogenous conditions within each host individual2.
Several studies have demonstrated that the colonization and accumulation of Streptococcus mutans is associated with dental caries in humans, since they are influenced by various factors in the oral cavity, such as nutrition and hygiene conditions of the host, salivary components, cleaning power and salivary flow and characteristics related with microbial virulence factors3. Streptococcus mutans is the key contributor to the formation of polysaccharide-based extracellular matrices in dental biofilms4. Glucosyltransferases synthesize exopolysaccharides, which are glucans that promote the accumulation of microorganisms at specific sites on dental surfaces, and glucosyltransferases become enzymatically active when exposed to dietary sucrose.
Streptococcus mutans is widely distributed not only among populations with a moderate-to-high incidence of caries5 but also among populations with little or no incidence of the disease6, which suggests that colonization by this microorganism alone is not sufficient for the development of caries5. Therefore, the presence of S. mutans in individuals with low caries experiences could be explained by differences in bacterial virulence factors or endogenous factors within the host populations. This microorganism is only one member of the indigenous oral biota that plays roles in "severe early childhood caries" (S-ECC). As most humans are host to S. mutans but not all carriers manifest dental caries, it has been suggested that certain strains of S. mutans associated with SECC could be genetically different from strains found in caries-free individuals7.
Molecular markers have been used to elucidate the genetic variability among S. mutans isolated from saliva and other sources. RAPD (Random Amplified Polymorphic DNA) markers have been most widely used for this purpose8-10. In addition, IGS sequencing (e.g., sequencing of intergenic regions between the 16S and 23S rRNA genes) has been used to taxonomy studies, while partial sequencing of the gene gtfB, which encodes the enzyme glucosyltransferase B, has been used to investigate enzymatic activity and virulence11-12.
In the present study, samples of S. mutans isolated from the saliva of children with varying histories of dental caries were characterized using morphological and biochemical markers, 16S-23S IGS sequencing, and RAPD markers. These isolates were further distinguished by sequencing specific regions of the gtfB gene to characterize the genetic variability at this locus and correlate this information with the colonization profiles of S. mutans observed in the children.
Material and methods
Case series and strains
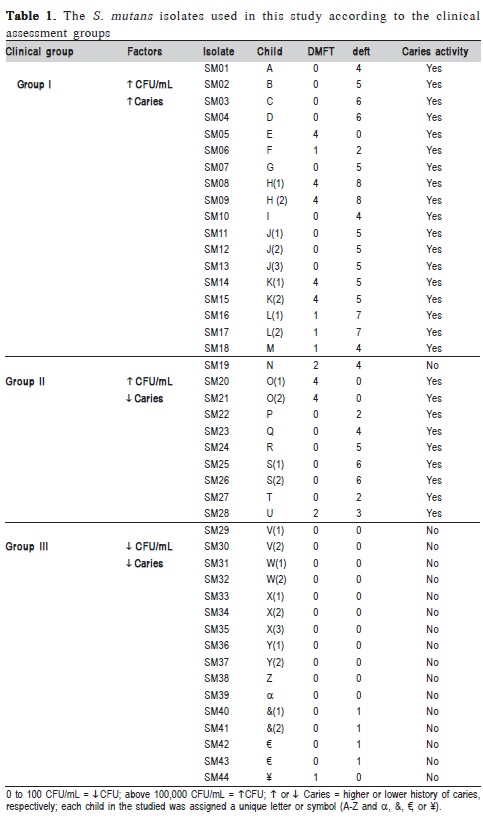
For the present study, it was selected 44 strains of Streptococcus mutans isolated from the saliva of 30 children aged 6-9 years, attending rural schools affiliated with the University Extension Program "Multidisciplinary strategy for the prevention and control of diseases manifesting clinically in childhood"13. The sample of children exhibiting a range of epidemiological profiles of dental caries was initially characterized according to the DMFT (the number of permanent decayed, missing, or filled teeth) and deft (deciduous teeth) metrics. The isolation of the strains and estimation of colony-forming units (CFU) per mL of saliva were performed using Mitis Salivarius Agar. Based on the biochemical evidence, as well as factors identified during the clinical interviews and examinations, the 44 S. mutans isolates were divided into three host groups (Table 1).
The study protocol was approved by the Research Ethics Committee of the Federal University of Paraná (UFPR), number 085.SI 048/04-06. Informed consent was provided by the children's legal representatives.
DNA extraction and RAPD analysis
DNA extraction was performed according to Moreira et al.14 (2010) by applying an ultrasound to a combination of silica and celite (2:1) in CTAB (cetyltrimethylammonium bromide). For the RAPD reactions, the following six oligonucleotide primers were used: OPA 2 (5' TGCCGAGCTG 3'), OPA 3 (5' AGTCAGCCAC 3), OPA 5 (5' AGGGGTCTTG 3'), OPA 8 (5' GTGACGTAGG 3'), OPA 9 (5' GGGTAACGCC 3') and OPA 13 (5' CAGCACCCAC 3'). The PCR was performed using the conditions as described previously9. The Streptococcus mutans strains ATCC 25175 and UA159 were included as reference samples during the amplifications, and the strains S. sobrinus ATCC 33478 and S. pyogenes ATCC 13540 were included as external control groups. The products resulting from the RAPD amplification were analyzed by electrophoresis through 1.6% agarose gels. An analysis of polymorphisms was performed using the non-weighed pair group method with arithmetic mean (UPGMA)15 (with an estimated Jaccard similarity coefficient) using the NTSYS 2.1 software program16. The consistency of the clustering was verified by bootstrap analysis with 10,000 re-samplings using the Bood 3.03 software program17; clusters exhibiting p values equal to or higher than 75% were considered consistent.
Amplification and sequencing of the 16S-23S intergenic region and gtfB gene
The 16S-23S intergenic region was amplified from the total DNA taken from the S. mutans isolates using primers 13BF (5' GTGAATACGTTCCCGGGCCT 3') and 6R (5' GGGTTYCCCCRTTCRGAAAT 3')18. In order to sequence a region of the gtfB gene, the following primers were used: GTFB-F (5' ACTTACACACTTTTCGGGTGGCTTGG 3') and GTFB-R (5' CAGTATAAGCGCCAGTTTCATC 3')19.
The sequencing reactions were composed as follows: 50 ng of PCR product purified with PEG (polyethylene glycol) according to Murphy et al.20 (2005); 125 μM each dNTP; 50 nM each primer; 0.5 μL Big-Dye Terminator V 3.1 Cycle Sequencing kit; 0.5 μL buffer and ultrapure water for a final volume of 10 μL. The amplification conditions were as follows: 1 min at 96 ºC, followed by 35 cycles at 96 ºC for 10 s and 50 ºC for 5 s with a final elongation step at 60 ºC for 4 min. Following lyophilization in a SpeedVac at 60 ºC for 40 min, the samples were sequenced using electrophoresis in an ABI Prism 377 Sequencer (Applied Biosystems). The sequences were analyzed and corrected using the Staden Package software program21 and aligned using a version of CLUSTAL-W22. In order to obtain the nucleotide and amino acid compositions and calculate the variable nucleotide distances and positions, we used the MEGA 3 software program23. Reference strains sequence data were taken from the GenBank.
Results
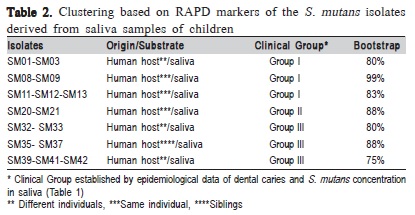
Forty-four S. mutans isolates were analyzed in this study. Based on the amplification profiles of the RAPD markers, polymorphisms were identified among the investigated isolates, and further analysis clustered the isolates into seven groups with bootstrap values above 75% (Table 2).
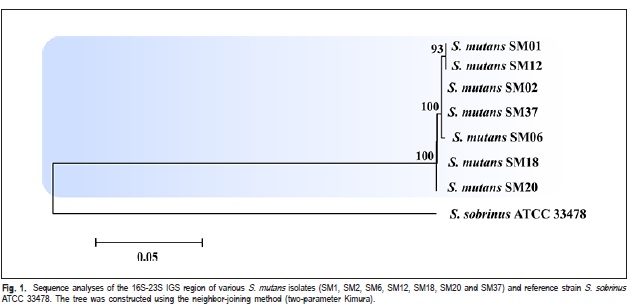
Thus, isolates from different genetic groups determined by RAPD markers, were characterized by intergenic regions (16S-23S) sequencing of rDNA and identified as S. mutans (Figure 1). Based on the analyzed sequences of intergenic region, it was verified the existence of genetic variability among these isolates. The isolates SM01, SM02, SM06, SM12 and SM37 exhibited a high degree of genetic similarity among them (Figure 1). The isolates SM01 and SM12 (Group I, Table 1) showed 100% of similarity. The Strain 33478 of S. sobrinus, which was included as an external control group, clustered at a distance from the experimental samples.
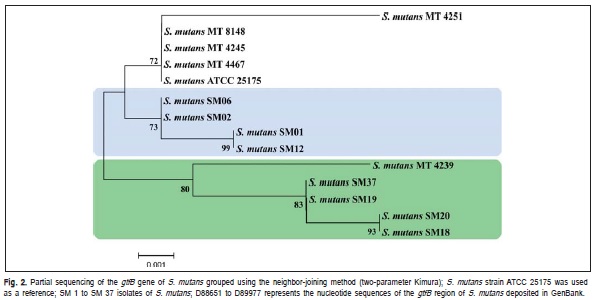
The characterization of isolates by partial sequencing of the gene encoding the enzyme glucosyltransferase B (GtfB>) was also done. As shown in Figure 2, isolates SM01, SM02, SM06 and SM12, originated from individuals with similar disease histories (Table 1), clustered in the same group; isolates SM01 and SM12 were found to be identical with respect to the analyzed sequences, although they originated from different individuals, but with the same patterns of clinical manifestation, i.e. high caries status (Table 1).
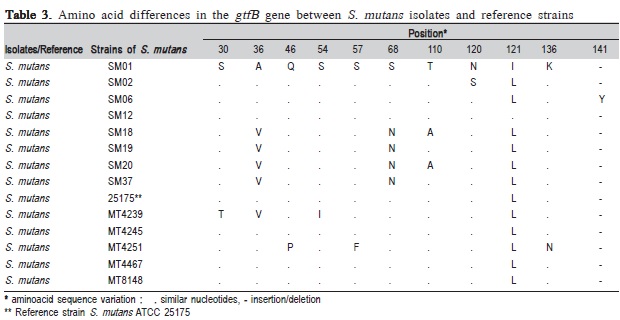
The analyzed isolates exhibited amino acid sequence variations, which we compared with the sequences of the reference strains as previously described by Fujiwara et al.11 (1998). The S. mutans reference strain (MT4239)11 clustered with isolates SM18, SM19, SM20 and SM37 (Figure 2), which were similar with respect to amino acid composition in the position 36 of the enzyme (Table 3).
Discussion
Among the 30 children evaluated in the study, 13 exhibited high concentrations of S. mutans in their saliva (> 105 CFU/mL) and had significant histories of disease (both DMFT and deft), whereas 17 children exhibited lower concentrations of S. mutans (<102 CFU/mL) and mostly had only mild histories of disease (Table 1). These data indicate a positive correlation between S. mutans concentrations in saliva and the clinical manifestation of caries. However, among the children exhibiting low levels of colonization, one group (Group II) had dental caries, despite having low concentrations of S. mutans in their saliva, which suggests that differences might also exist with respect to the virulence of the isolates.
According to the polymorphisms identified by the RAPD markers, it was identified a relation between genetic groups of S. mutans and epidemiological data of disease (Table 2). It was also verified the clustering of isolates from different individuals, supported by high bootstrap values, suggesting that isolates can be transmitted among individuals. Such results correspond to what has been previously reported by Domejean et al.8 (2010) who used arbitrary primers and verified the presence of S. mutans corresponding genotypes, proving the horizontal transmission of bacteria among schoolchildren, aging between 5 and 6 years old. Furthermore, according to the results presented in Table 2, it was verified two isolates grouping proceeding from the same individual (e.g., SM11- 13 and SM20-21).
Furthermore, these results (Table 2) demonstrate the diversity among isolates, correlated with patterns of variation in the clinical manifestation of the disease, suggesting that each individual may exhibit unique patterns of colonization by S. mutans that could be due host's endogenous factors, since these individuals had a common pattern of diet, characterized by frequent consumption of refined carbohydrates.
According to Kreth et al.24 (2005), human mucosal surfaces are colonized by large numbers of bacterial species; these populations exist in a state of homeostasis and play an important role in protecting the host against invasion by exogenous pathogens. However, when this homeostasis is disrupted, indigenous flora can cause pathologies, such as dental caries and periodontal disease. Additionally, high colonization levels and diverse range of S. mutans genotypes could be a consequence of frequent consumption of fermentable carbohydrates, and it is possible that the simultaneous action of multiple S. mutans strains with different cariogenic potentials could increase the risk of developing dental caries25. In addition, when investigating the children's oral microbiota, Crielaard et al.26 (2011), showed that the salivary microbiota of children aged 3 to 18 years are still maturing and that multiple colonization events can occur during this period.
The isolates from different genetic groups determined by RAPD markers (Table 2), were characterized by intergenic regions (16S-23S) sequencing of rDNA, and it was verified the existence of genetic variability among these isolates. The isolates SM01 and SM12 (Group I, Table 1), which were originated from different children, showed 100% similarity. This result suggested the transmissibility of S. mutans among schoolchildren (Figure 1). This way, it can notice that the variability among the isolates, detected by the IGS sequencing (Figure 1), is not related to the virulence, but may indicate the route of transmissibility.
According to the literature, the activity of glucosyltransferase B enzyme seems to be directly related to the adherence of S. mutans to dental enamel4-27. Upon partial sequencing of the gtfB gene, five different genotypes were identified among the analyzed isolates. The nucleotide sequences differences were predicted to result in amino acid variations that could influence the patterns of virulence among the isolates. Figure 2 shows the grouping of the isolates originating in children with varying disease histories, which were clustered according to their genetic similarity identified by partial sequencing of the gtfB gene.
The finding of isolates SM01 and SM12 clustered in the same group, and originated from different individuals with the same patterns of clinical manifestation, i.e. high caries status (Table 1), suggests the existence of a similar virulence potential for these two isolates.
The genetic similarity of isolates SM20 and SM18 proceeding from individuals with different patterns of colonization, as evidenced by differences in saliva CFU/mL counts (Clinical Groups I and II, Table 1) and high disease prevalence, also suggested a similar pattern of virulence in these isolates. The isolates SM19 and SM37, proceeding from children with low concentration of S. mutans and with different caries historic (Clinical Group II and III, Table 1), was grouped with the latter, indicating that both exogenous and endogenous host factors also might influence disease development.
Based on the results of amino acid sequence variations described in Table 3, amino acid positions ranging from 30 to 141 appeared to be susceptible to variation of S. mutans GtfB gene. According to the literature, the presence of polymorphisms in glucosyltransferase (gfts) genes expressed by different strains of S. mutans is likely to be associated with variations in specific enzymatic activity28. Alterations by the site-directed mutagenesis of a single amino acid in the catalytic domain of Gtfs are sufficient to considerably change the enzymatic activity of this protein, leading to differences in dental surface adherence patterns for the investigated strains of S. mutans.
It is important to emphasize that the isolate SM06 exhibited an insertion of tyrosine at position 141, which differed from the rest of the isolates (Table 1). Such modification would be likely to influence enzymatic activity, as reports indicate that this is the most highly conserved position in GtfB due to its location within the catalytic domain29. This hypothesis is consistent with the observed pattern of virulence for isolate SM06, which was originated in an individual with a considerable clinical history of disease.
In addition, glucosyltransferases produced by S. mutans are also known to play an important role in virulence due to their effects on dental biofilms, according to phylogenetic analysis of glucosyltransferases and implications for the coevolution of mutants streptococci with their mammalian host, as reported by Argimón et al.30 (2013).
Glucosyltransferases can be adsorbed directly onto enamel and catalyze the synthesis of glucans in situ ; this promotes sites of avid colonization by microorganisms and helps to create an insoluble matrix of dental bacterial plaque27, which might also favor the development of disease.
The occurrence of groups containing children with a high clinical manifestation of disease and low indices of S. mutans in their saliva indicates that in addition to the endogenous and exogenous factors present in each individual, the presence of genetic variations must also be taken into account when attempting to estimate the virulence of wild biotypes of S. mutans.
In conclusion, this study demonstrates that partial sequencing of the gene encoding the enzyme glucosyltransferase B can be a useful tool for elucidating the virulence patterns of S. mutans because amino acid variations are likely to justify the differences in colonization pattern of these strains, which could reflect a biological risk of development the dental caries in these children.
Acknowledgements
The authors are grateful for the support of the UFPR, CAPES (Federal Agency of Support and Evaluation of Postgraduate Education/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPQ and the Araucaria Foundation.
References
1. Berkowitz RJ. Mutans streptococci: acquisition and transmission. Pediatr Dent. 2006; 28: 106-9. [ Links ]
2. Phattarataratip E, Olson B, Broffitt B, Qian F, Brogden KA, Drake DR, et al. Streptococcus mutans strains recovered from caries-active or cariesfree individuals differ in sensitivity to host antimicrobial peptides. Mol Oral Microbiol. 2011; 26: 187-99.
3. Kamiya RU, Taiete T, Gonçalves RB. Mutacins of Streptococcus mutans. Braz J Microbiol. 2011; 42: 1248-58.
4. Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010; 192: 3024-32.
5. Beigton D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent Oral Epidemiol. 2005; 33: 248-55.
6. Matee MIN, Mikx FHM, De Soet JS, Maselle SY, De Graff J, Van Palenstein Helderman WH. Mutans streptococci in caries-active and caries Braz J Oral Sci. 13(3):235-241 free infants in Tanzania. Oral Microbiol Immunol. 1993; 8: 322-4.
7. Saxena D, Caufield PW, Li Y, Brown S, Song J, Norman R. Genetic classification of severe early childhood caries by use of subtracted DNA fragments from Streptococcus mutans. J Clin Microbiol. 2008; 46: 2868-73.
8. Domejean S, Zhan L, DenBesten PK, Stamper J, Boyce WT, Featherstone JD. Horizontal transmission of Mutans streptococci in children. J Dent Res. 2010; 89: 51-5.
9. Moreira M, Vicente VA, Glienke C. Genetic Variability of Streptococcus mutans isolated from low-income families, as shown by RAPD markers, Braz J Microbiol. 2007; 38: 729-35.
10. Spolidorio DMP, Hofling JF, Pizzolito AC, Rosa EA, Negrini TC, Spolidoro LC. Genetic polymorphism of Streptococcus mutans in brazilian family members. Braz J Microbiol. 2003; 34: 213-7.
11. Fujiwara T, Terao Y, Hoshino T, Kawabata S, Ooshima T, Sobue S, et al. Molecular analyses of glucosyltransferase genes among strains of Streptococcus mutans. FEMS Microbiol Lett. 1998; 161: 331-6.
12. Hoshino T, Kawaguchi M, Shimizu N, Hoshino N, Ooshima T, Fujiwara T. PCR detection and identification of oral streptococci in saliva samples using gtf genes. Diagn Microbiol Infect Dis. 2004; 48: 195-9.
13. Vicente VA, Moreira M, Zardo EL, Braga SF, Costa AO, Neiva IF, et al. Multidisciplinary strategy for the prevention and control of diseases clinically manifesting in childhood. RGO. 2011; 59: 591-7.
14. Moreira M, Noschang J, Neiva IF, Carvalho Y, Higuti IH, Vicente VA. Methodological Variations in the isolation of genomic DNA from Streptococcus bacteria. Braz Arch Biol Technol. 2010; 53: 845-9.
15. Meyer D. Human evolutionary trees: a discussion on phylogenetic interference. Monographs series. 1993; 3.
16. Rohlf FJ. NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software. New York; 2000. 98p.
17. Coelho ASG. Assessment of dendrograms based on estimates of the genetic distances/similarities by means of the bootstraps procedure. BOOD 3.03. Goiânia, GO: Department of General Biology, Institute of Biological Sciences, Federal University of Goiás; 2005.
18. Chen CC, Teng LJ, Chang TC. Identification of clinically relevant viridans group streptococci by sequence analysis of the 16S-23S ribosomal DNA spacer region. J Clin Microbiol. 2004; 42: 2651-7.
19. Oho T, Yamashita Y, Shimazaki Y, Kushiyama M, Koga T. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol Immunol. 2000; 15: 258-62.
20. Murphy KM, Berg KD, Eshleman JR. Sequencing of genomic DNA by combined amplification and cycle sequencing reaction. Clin Chem. 2005; 51: 35-9.
21. Staden R, Judge DP, Bonfield JK. Sequence assembly and finishing methods. Methods Biochem Anal. 2001; 43: 303-22.
22. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22: 4673-80.
23. Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004; 5: 150-63.
24. Kreth J, Merrit J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus Sanguinis in the dental biofilm. J Bacteriol. 2005; 187: 7193-203.
25. Alaluusua S, Matto J, Gronroos, L, Innila, S, Torkko, H, Asikainem, S, et al. Oral colonization by more than one clonal type of Mutans streptococci> in children with nursing-bottle dental caries. Arch Oral Biol. 1996; 41: 167-73.
26. Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011; 4: 22.
27. Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011; 45: 69-86.
28. Chia JS, Hsu TY, Teng LJ, Chen JY, Hahn LJ, Yang CS. Glucosyltransferase gene polymorphism among Streptococcus mutans strains. Infect Immun. 1991; 59: 1656-60.
29. Fujiwara T, Hoshino T, Ooshima T, Hamada S. Differential and quantitative analyses of mRNA expression of glucosyltransferases from Streptococcus mutans MT8148. J Dent Res. 2002; 81: 109-13.
30. Argimón S, Alekseyenko AV, DeSalle R, Caufield PW. Phylogenetic analysis of glucosyltransferases and implications for the coevolution of Mutans streptococci with their mammalian hosts. PLos ONE 2013; 8: e56305.
 Correspondence:
Correspondence:
Vânia Aparecida Vicente
Universidade Federal do Paraná - UFPR
Area of Biological Sciences
Caixa Postal: 19020
CEP 81.531-980 - Curitiba, PR, Brasil
E-mail: vicente@ufpr.br
Received for publication: August 22, 2014
Accepted: September 16, 2014