Serviços Personalizados
Artigo
Links relacionados
Compartilhar
Brazilian Journal of Oral Sciences
versão On-line ISSN 1677-3225
Braz. J. Oral Sci. vol.14 no.2 Piracicaba Abr./Jun. 2015
ORIGINAL ARTICLE
Expression analysis of Notch signaling pathway molecules in SHED cultured in keratinocyte growth medium
Siti Aisyah Mohd TahaI;Joanne Koh Su LingI; Nur Izyan Binti AzmiI; Thirumulu Ponnuraj KannankI,II; Ahmad AzlinaI; Khairani Idah MokhtarIII
I Universiti Sains Malaysia, School of Dental Sciences, Kota Bharu, Kelantan, Malaysia
II Universiti Sains Malaysia, School of Medical Sciences, Human Genome Centre, Kota Bharu, Kelantan, Malaysia
III International Islamic Universiti Malaysia, Kulliyah of Dentistry, Kuantan, Pahang, Malaysia
Abstract
Aim: To detect the expression of molecules associated with Notch signaling pathway in stem cells from human exfoliated deciduous teeth (SHED) cultured in specific differentiation medium, namely, keratinocyte growth medium (KGM). Methods: RNA was extracted from SHED harvested on day 1, 3 and 7. RNA was reverse-transcribed to obtain the cDNA and then proceeded with PCR using specific primers for the Notch signaling pathway molecules (Notch1, Jagged-1, Jagged-2 and, Hes1) as well as stem cell marker (Nanog). PCR products were electrophoresed on a 2% agarose gel and stained with SYBR green. Results: Notch-1 was highly expressed in SHED cultured in KGM and showed increase in density as the days progressed, while Jagged-1 showed a decrease. Jagged-2 on the other hand, showed a slight increase on day 3 followed by a decrease on day 7. However, Hes-1 was not expressed in SHED cultured in KGM. Nanog showed expression only on day 3 and gradually increased in expression on day 7. Conclusions: Notch signaling pathway associated molecules; Notch-1, Jagged-1, Jagged-2, and stem cell marker Nanog are expressed in SHED cultured in KGM which may be involved in the differentiation into epithelial-like cells in human dental pulp tissues.
Keywords: receptors, notch; gene expression; stem cells; tooth, deciduous; culture media.
Introduction
Stem cells from human exfoliated deciduous teeth (SHED) are multipotent stem cells derived from the pulp tissues of extracted deciduous teeth1. SHED has the ability to be differentiated to specific cell lineages such as odontoblasts and osteoblasts as well as epithelial like cells. SHED was able to differentiate into epithelial like cells when cultured in keratinocyte growth medium (KGM)2. Since the Notch signaling pathway molecules play an important role in differentiation of epithelial cells, it is important to identify the presence of notch signaling molecules in SHED during the process of cell differentiation.
The Notch signaling pathway provides important intercellular signaling mechanisms essential for cell fate specification and it regulates differentiation and proliferation of stem or progenitor cells by para-inducing effects3-4. The core components of a Notch signaling pathway involves three different molecules; the DSL-type ligand, a Notch-type receptor and a transcription factor of the CSL family (CBF1; C promoter binding factor/ Suppressor of Hairless/ lag- 1). Notch signaling pathway is also involved in the regulation of epithelial cell differentiation in various tissues5-6.
The aim of this study was to detect the expression of molecules associated with Notch signallng pathway in SHED cultured in specific differentiation medium, namely, KGM. Knowledge on the expression analysis of Notch signaling pathway molecules in SHED cultured in KGM could highlight its involvement in controlling the biological activity of these stem cells, particularly during odontogenesis and other developmental process.
Material and methods
SHED culture
Stem cells from human exfoliated deciduous teeth (SHED) (ALLCells, Alameda, CA, USA) were employed in the current study. SHED was cultured and maintained in T25 cm2 culture flask using Minimum Medium Alpha (α-MEM) from Gibco, USA. The medium was changed 3 days after culture and were sub-cultured once they reached 70% confluence. At each passage, the cells were counted, photographed using an inverted phase-contrast microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA) and cyro-preserved in cyrovial tube for further culture and later for RNA analysis. The cryopreserved SHED was subcultured for 2 passages in α-MEM. After washing, 1x10ˆ6 cells were seeded and cultured in KGM (Lonza Group Ltd., Basel, Switzerland). Total RNA was extracted from the cells cultured in KGM after 1, 3 and 7 days.
RNA extraction
The total RNA was extracted from the cells using RNA extraction kit, RNeasy Mini Kit (Qiagen Inc., Valencia, CA USA) following the protocol provided by the manufacturer.
cDNA synthesis
Reverse-transcriptase enzyme was used to convert extracted RNA into cDNA using reverse transcriptase cDNA synthesis kit (MMLV RT 1st-Strand cDNA synthesis kit, Korea) following the protocol provided by the manufacturer. 8.23 microlit (200 ng) of total RNA was used for cDNA synthesis using Oligo(dT)21 Primer. The synthesized cDNA was stored at -20 oC and was used as a template for PCR technique using specific primer pairs. The quality of cDNA was observed by running the sample on a 2% agarose gel.
Polymerase chain reaction (PCR)
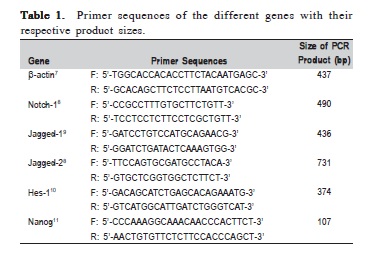
The synthesized cDNA was used for PCR amplification using specific pair of primers for the respective genes, Notch1, Jagged-1, Jagged-2 ligands and Hes-1 transcription factor genes and Nanog, stem cell marker gene. The sequence of the primers with their references and respective product sizes are presented in Table 1. The house-keeping gene used in this study was β-actin. The PCR master mix was made in a total volume of 50 μl of reaction mixture containing 1 μl (200 ng) of cDNA template, 1 μl (10 μM) of each forward and reverse primers, 25 μl of PCR ready-to-use mixture (MyTaq HS Mix 2x, Bioline, London, UK) and 22 μl of distilled water.
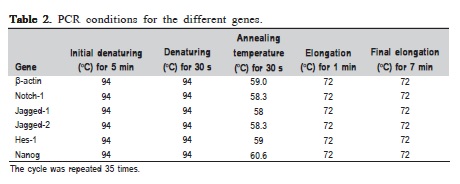
The PCR conditions used were similar for all genes but with different annealing temperatures using PCR machine (C1000 Thermal Cycler, Biorad, Hercules, CA, USA) as shown in Table 2. Two μl of the PCR products were electrophoresed on 2% agarose gel in LB buffer at 100V (Power Pac HC, Biorad) and visualized under UV after SYBR green staining. The gel was photographed under UV light using digital image analyzer (GEL Doc XR, Biorad). Appropriate product size of the specific genes analyzed was indicated by the production of a discrete single band on the 2% agarose gel. The experiments were run in triplicates and the average density value (ADV) of the PCR products for each gene was calculated.
Statistical analysis
The data obtained were analyzed using the Kruskal- Wallis rank test.
Results
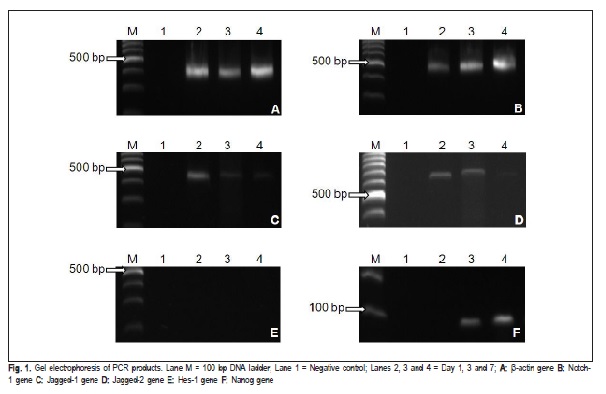
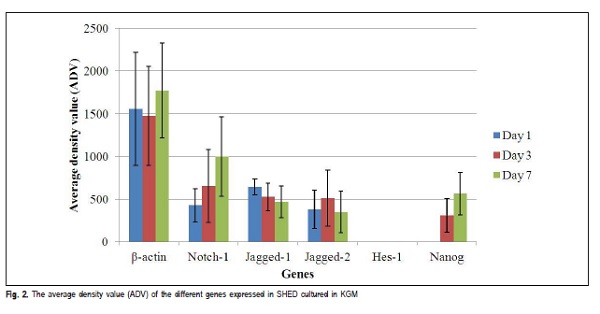
The house keeping gene β-actin expressed with a band size of 437 bp in all the samples. The product size for β- actin of SHED cultured in KGM for day 1, 3 and 7 are shown in Figure 1A and the ADV is presented in Figure 2.
The samples analyzed for the expression of Notch-1 receptor in this study cultured in KGM showed expression of Notch-1, which increased in expression as the days progressed from day 1 to, 3 and 7. This is shown in Figure 1B with the ADV in Figure 2.
The samples were tested for the expression of Jagged-1. All the samples showed expression of Jagged-1 , which decreased with increase in days. The expression for Jagged- 1 of SHED cultured in KGM for day 1, 3 and 7 is shown in Figure 1C and the ADV is presented in Figure 2.
The samples tested for the expression of Jagged-2 in this study showed expression of Jagged-2 on all the days but showed a different pattern of expression. The expression increased from day 1 to day 3 and then followed by a decrease on day 7. The expression of Jagged-2 of SHED cultured in KGM for day 1, 3 and 7 are shown in Figure 1D and the ADV is presented in Figure 2.
The samples were also analyzed for the expression of Hes-1 transcription factor. In this study, all samples did not show any expression of Hes-1 in SHED cultured in KGM for day 1, 3 and 7 as shown in Figure 1E.
The expression analysis for stem cell marker, Nanog was also performed. Nanog was not detected on day 1 but was expressed at day 3 and 7 as shown in Figure 1F. The average density value (ADV) for all analyzed PCR products is presented in Figure 2.
Discussion
To evolve specific developmental programs, Notch signals control cells that respond to intrinsic or extrinsic developmental cues. Development of morphology of organs and their evolution are affected by differentiation, proliferation, and apoptotic programs via the Notch activity4.
The DSL-type ligand, a Notch-type receptor and a transcription factor of the CSL family are the three different molecules which are the main components of Notch signaling pathway3,12,. When the ligands on neighboring cells bind to Notch receptor on one cell, it will generate an active Notch intracellular fragment (NIC) or Notch intracellular domain (NICD). In order to trigger the transcription of Notch target genes, the active NICD will be released into the cytoplasm before it translocates into the nucleus12.
The main functions of Notch signaling are differentiation of odontoblasts and osteoblasts, cusp pattern formation, tooth roots generation and calcification of tooth hard tissue. The Notch signaling can also be triggered in dental stem cells of the pulp to differentiate it into odontoblast, and forming fresh dentin tissue after tooth eruption3. Additionally, Notch signaling has been shown to be required for epithelial stem cell survival and amelogenesis13.
SHED is a mesenchymal stem cell derived from the pulp tissues of extracted deciduous teeth and is found to be a population of highly proliferative, clonogenic cells capable of differentiating into a variety of cell types including neural cells, adipocytes, and odontoblasts1. As it is deemed that the Notch signaling pathway molecules have a role to play in the differentiation of epithelial cells, it would be appropriate to investigate the notch signaling molecules in SHED during the process of cell differentiation.
The Notch receptor family is mainly expressed in tooth germ during the early development stage, throughout the dental epithelium and during the differentiation stage in the stratum intermedium, gradually extending to the pulp mesenchyme14.
In addition to mediating tissue-tissue interactions, growth factor signaling also participates in mediating cellcell interactions within epithelial and mesenchymal tissues during organogenesis15. This short-range signaling between cells is usually accomplished by one cell possessing a transmembrane receptor and the neighboring cell possessing a membrane-bound ligand. Notch signaling is one such system implicated in tooth morphogenesis14-15. Notch-1 gene encodes a member of the Notch family. Members of this Type 1 transmembrane protein family share structural characteristics including an extracellular domain consisting of multiple epidermal growth factor-like (EGF) repeats, and an intracellular domain consisting of multiple, different domain types16. In this study, Notch-1 gene was detected in SHED cultured in KGM, with marked increase from day 1 to day 3 and day 7 but it was not statistically significant (p>0.05). This shows that Notch-1 is being expressed, suggesting the presence of Notch signaling pathway receptor. This is parallel to the study shown by Mitsiadis and his co-workers17 where the expression of Notch-1, -2, and -3 was detected in the dental epithelium from the initiation stage to the later differentiation stage.
Jagged transmembrane proteins are ligands to the Notch receptors. Intimate cell-cell contacts aid in binding of the membrane-bound ligands to the Notch receptor and triggers the release of the cytoplasmic domain of Notch that functions as a transcription factor in the nucleus. The expression of several Notch ligands, one of them Jagged-1 , has been detected in developing teeth18. This study showed presence of Jagged-1 gene in the SHED cultured in KGM. This showed that there is a membrane-bound ligand to Notch receptors, indicating the function played by Notch signaling pathway during the differentiation process. According to Zhang et al. the upregulation of Jagged-1 indicated that SHED might differentiate into epithelial-like cells and inhibit to differentiate into odontoblast-like cells19. In the current study, though the expression of Jagged-1 showed a declining trend, yet, it was not statistically significant (p>0.05).
Jagged-2 is another ligand to Notch receptors. Jagged-2 gene provides instructions for making a protein, which is part of JAK/STAT pathway that promotes the growth and proliferation of cells. This pathway transmits chemical signals from extracellular to the cell's nucleus20.
In our study, it was found that Jagged-2 was expressed at day 1, increased at day 3 but eventually, the level of expression decreased by day 7, where, day 7 had the lowest level of expression of this gene. Though there was fluctuation in the expression of Jagged-2, yet, there was no statistical significance as the p>0.05. It was down regulated as compared to β-actin. This showed that Jagged-2 is also involved in the differentiation process of SHED into epithelial-like cells through Notch signaling pathway. Interestingly, the current study also showed that both ligands; Jagged-1 and Jagged-2 were down regulated at day 7 while Notch-1 expression was up regulated at this point. This may suggest that as the process of differentiation continues, the expression of ligands start to subside. In our study, the Hes-1 gene showed no expression at all throughout the process. Hes-1 is activated transcriptionally by the Notch signaling pathway during Notch-mediated lateral inhibition21. Hes-1 is one of the downstream transcription factors involved in Notch signaling pathway and has been shown to be important in regulating the maintenance of the progenitor or stem cells. There are seven members in the Hes family (Hes1-7), thus suggesting that other members of Hes family might be involved in this process of controlling the stem cell differentiation22-23. Though Hes-1 is a downstream target of notch signaling, may be it is not expressed in SHED cultured in KGM.
Nanog is the protein coding gene that acts as a transcription regulator involving in inner cell mass and embryonic stem (ES) cell proliferation and self-renewal10-24. In our study, Nanog started showing the expression at day 3 and subsequently increased in density at day 7, which shows the stemness of this SHED. However, there was no statistical significance seen (p>0.05).
This study showed that the Notch signaling pathway associated molecules, Notch-1, Jagged-1, Jagged-2, and stem cell marker Nanog are expressed in SHED cultured in KGM. This indicates that these molecules may be involved in the differentiation into epithelial-like cells in human dental pulp tissues.
Acknowledgements
The authors would like to acknowledge the staff of Craniofacial Science Laboratory, School of Dental Sciences, Universiti Sains Malaysia for their technical support. This study was supported by Universiti Sains Malaysia Research University Grant (1001/PPSG/813077).
References
1. Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003; 100: 5807-12. [ Links ]
2. Nam H, Lee G. Identification of novel epithelial stem cell-like in human deciduous dental pulp. Biochem Biophys Res Commun. 2009; 386: 135-9.
3. Cai X, Gong P, Huang Y, Lin Y. Notch signalling pathway in tooth development in adult dental cells. Cell Prolif. 2011; 44: 495-507.
4. Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: Molecular basis of cell context dependency. Eur J Cell Biol. 2011; 90: 572-81.
5. Leong KG, Karsan A. Recent insights into the role of Notch signalling in tumorigenesis. Blood. 2006; 107: 2223-33.
6. VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012; 139: 488-97.
7. Karaoz E, Dogan BN, Aksoy A, Gacar G, Akyuz S, Ayhan S, et al. Isolation and in vitro characterisation of dental pulp stem cells from natal teeth. Histochem Cell Biol. 2010; 133: 195-12.
8. Zhang XP, Zheng G, Zou L, Liu HL, Lou LH, Zhou P, et al. Notch activation promotes cell proliferation and the formation of neural stem cell-like colonies in human glioma cells. Mol Cell Biochem. 2008; 307: 101-8.
9. Tachikawa Y, Matsushima T, Abe Y, Sakano S, Yamamoto M, Nishimura J, et al. Pivotal role of Notch signalling in regulation of erythroid maturation and proliferation. Eur J Haematol. 2006; 77: 273-81.
10. Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003; 113: 643-55.
11. Park SW, Lim HY, Do HJ, Sung B, Huh SH, Uhm SJ, et al. Regulation of human growth and differentiation factor 3 genes expression by NANOG in human embryonic carcinoma NCCIT cells. FEBS letter. 2012; 586: 3529-35.
12. Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity. 2010; 32: 14-27.
13. Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010; 80: 241-8
14. Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010; 137: 3025-35.
15. Thesleff I, Mikkola M. The role of growth factors in tooth development. Int Rev Cytol. 2002; 217: 93-135.
16. NOTCH1 [Homo sapiens (human)] [Internet]. HUGO Gene Nomenclature Committee – HGNC. Bethesda: National Center for Biotechnology Information, U.S. National Library of Medicine [cited 2014 Oct 15]. Available from: http://www.ncbi.nlm.nih.gov/gene/4851.
17. Mitsiadis TA, Feki A, Papaccio G, Catón J. Dental pulp stem cells, niches, and notch signaling in tooth injury. Adv Dent Res. 2011; 23: 275-9.
18. Mitsiadis T, Henrique D, Thesleff I, Lendahl U. Mouse Serrate-1 (Jagged-1) expression in the developing tooth is regulated by epithelial-mesenchymal interactions and fibroblast growth factor-4. Development. 1997; 124: 1473-83.
19. Zhang C, Chang J, Sonoyama W, Shi S, Wang CY. Inhibition of human dental pulp stem cell differentiation by Notch signaling. J Dent Res. 2008; 87: 250-5.
20. Genetics Home Reference. Your Guide to Understanding Genetic Conditions [Internet]. Bethesda: National Center for Biotechnology Information, U.S. National Library of Medicine [cited 2014 Oct 15]. Available from: http://ghr.nlm.nih.gov/gene/JAK2.
21. Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995; 377: 355-8.
22. Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007; 134: 1243-51.
23. Kobayashi T, Kageyama R. Hes1 regulates embryonic stem cell differentiation by suppressing Notch signaling. Genes Cells. 2010; 15: 689-98.
24. Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003; 113: 631-42.
 Correspondence:
Correspondence:
Thirumulu Ponnuraj Kannan
School of Dental Sciences, Universiti Sains Malaysia
Health Campus 16150
Kota Bharu, Kelantan, Malaysia
E-mail: kannan@usm.my
Received for publication: April 09, 2015
Accepted: June 09, 2015