Serviços Personalizados
Artigo
Links relacionados
Compartilhar
RGO.Revista Gaúcha de Odontologia (Online)
versão On-line ISSN 1981-8637
RGO, Rev. gaúch. odontol. (Online) vol.58 no.2 Porto Alegre Abr./Jun. 2010
ORIGINAL ORIGINAL
Polymerase chain reaction technique as a diagnostic tool for bacterial detection in root canals of cleft lip and palate patients
Técnica de Reação em Cadeia da Polimerase como ferramenta de identificação da microbiota de canais radiculares em pacientes com fissuras labiopalatinas
Tatiana Cristina Silveira PereiraI; Thais Marchini OliveiraII, *; Vivien Thiemy SakaiII; Thiago José DionísioII; Renata Pardini HussneI; Celso Kenji NishiyamaI; Maria Aparecida Andrade Moreira MachadoII; Carlos Ferreira SantosII
IUniversidade de São Paulo, Hospital de Reabilitação de Anomalias Craniofaciais. Bauru, SP, Brasil
IIUniversidade de São Paulo, Faculdade de Odontologia. Alameda Dr. Octávio Pinheiro Brisolla, 9-75, 17012-901, Bauru, SP, Brasil
ABSTRACT
OBJETIVE: Polymerase chain reaction is the most sensitive of all microbiological methods for the detection of microorganisms. It consists of enzymatic amplification of DNA. PCR is faster, much more sensitive and more accurate than the culture method. This study investigated the occurrence of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in the root canals of cleft lip and palate patients.
METHODS: Samples were collected from 24 root canals followed by polymerase chain reaction.
RESULTS: A. actinomycetemcomitans was detected in 12.5% and P. gingivalis in 8.3% of the investigated root canals. The method proposed in this study was highly sensitive and specific for the direct detection of microorganisms in root canal samples.
CONCLUSION: This new molecular-based dentistry provides useful information for clarifying the etiology of root canal microbiota and for developing new strategies for endodontic diagnosis and treatment.
Indexing terms: cleft lip; cleft palate; dental pulp cavity; polymerase chain reaction.
RESUMO
OBJETIVO: A Reação em Cadeia da Polimerase é um método com alta sensibilidade e especificidade quando comparado com alguns métodos microbiológicos convencionais. Baseia-se na amplificação enzimática de uma sequência especifica de DNA, visando à produção de milhões de cópias desta sequência em um tubo de ensaio. Desta forma, recentemente as técnicas de biologia molecular têm sido usadas em endodontia pela sua rapidez e eficácia. Portanto, este estudo avaliou por meio da técnica de Reação em Cadeia da Polimerase a presença dos micro-organismos Actinobacillus actinomycetemcomitans e Porphyromonas gingivalis de canais radiculares em pacientes com fissuras lábio-palatais.
MÉTODOS: Foram coletadas amostras de 24 canais radiculares e realizada a técnica de Reação em Cadeia da Polimerase.
RESULTADOS: Das amostras estudadas, 12,5% mostraram resultado positivo para A. actinomycetemcomitans e 8,3% para P. gingivalis. O método proposto neste estudo foi altamente sensível e específico na detecção direta de amostras clínicas.
CONCLUSÃO: A odontologia com base molecular fornece informações úteis para esclarecer a etiologia da microbiota de canais radiculares e o desenvolvimento de novas estratégias de diagnóstico e tratamento endodôntico.
Termos de indexação: fenda labial; fissura palatina; cavidade pulpar; reação em cadeia da polimerase.
INTRODUCTION
Cultures have traditionally been used as reference for the assessment of microbiota associated with various infectious diseases, including those of endodontic origin1-6. Species identification is routinely done with agar plates and highly diluted samples, which may not correctly represent the sample in question. Therefore, nonviable or uncultivable bacteria cannot be isolated by culture methods. Recently, scientists have questioned if cultures are indeed the reference method for bacterial identification1,3-4,7.
A new era has been heralded for diagnosis microbiology. Microorganism detection methods that detect microbial DNA rather than the microorganisms themselves have been introduced in both research and clinical laboratories. These methods are revolutionizing the knowledge of infectious diseases by allowing effective and rapid diagnosis of many diseases8. Identifying and understanding both the etiologic factors and the pathophysiology of the disease process comprise the basis of any clinical treatment. This knowledge allows the development of rational solutions to deal with the problem. Since endodontic diseases are primarily of infectious etiology, finding the microbial species involved in the pathogenesis of these diseases is of utmost importance9.
Molecular methods have been used to detect microorganisms that are impossible or difficult to culture. Polymerase chain reaction (PCR) has the highest sensitivity of all the microbiological methods for the detection of microorganisms10. PCR is an in vitro method for replicating specific sequences of DNA. Starting from a very low quantity of DNA, as few as just one bacterial cell, PCR amplifies a billion-fold a specific known sequence of microbial DNA and allows its detection by electrophoresis6. Since PCR is highly sensitive and able to detect uncultivable microorganisms, it has potential applicability in endodontic research and diagnosis.
Root canal therapy success depends on complete eradication of the microflora within the root canal system. A great deal of research is needed to identify and define the role of the pathogens that are involved in the pathogenesis of periradicular diseases. For instance, Tannerella forsythensis, Treponema denticola, other Treponema species, Dialister pneumosites, and Prevotella tannerae were detected in infected root canals for the first time with PCR and were found to be highly prevalent. Moreover, other bacterial species, such as Porphyromonas endodontalis, Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and some Eubacterium spp. have been reported in endodontic infections more frequently when PCR is used than when cultures are used for such investigations 1,4-6,9.
The purpose of this study was to assess the prevalence of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in root canals of cleft lip and palate patients using polymerase chain reaction.
METHODS
Samples were collected from root canals of 24 teeth of adult patients admitted to our institution for pain management of endodontic origin. Selection was based on clinical and radiographic examination. All teeth presented carious lesions; initially, they were cleansed with pumice and isolated with a rubber dam. The tooth, the clamp, and the rubber dam were cleansed with 3% hydrogen peroxide and then disinfected with a 2.5% sodium hypochlorite solution. After completion of the access preparations, the operative field, including the pulp chamber, was swabbed with 2.5% sodium hypochlorite. The root canal was dried with paper point. Samples were initially collected by means of #15 K-type file with the handle cut off. The file was introduced to a level approximately 1mm short of the tooth apex and a discrete filing motion was applied. Two sequential paper points were placed on the same level and used to soak up the fluid in the canal. Each paper point was retained in position for 1 mm. The cut file and the two paper points were then transferred to a sterile microcentrifuge tube containing 0.5 mL of sterile water, and frozen at -20º C for later analysis.
For DNA extraction, microcentrifuge tubes were thawed and centrifuged at 10,000 X g for 5 minutes at 4oC. The supernatant was discarded and the resulting pellet was washed 2 more times with 1 ml of sterile water. The pellet was reconstituted with 100 µL of water and 100 µL of InstaGene Matrix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and incubated at 56o C for 30 minutes. The samples were then vortexed and boiled for 10 minutes. After centrifugation to remove unbroken cells and large debris (10,000 X g for 3 minutes), the supernatant was analyzed by PCR.
A total of 50 µL of PCR reaction mixture contained 10 µL of DNA sample, 5 µL of 10 X PCR buffer, 1.25 unit of Taq DNA polymerase, 0.2 mM each of deoxyribonucleotides, 1.0 µM of each primer, and 1.0 mM of MgCl2 for A. actinomycetemcomitans or 1.5 mM of MgCl2 for P. gingivalis. As positive controls, isolated DNA from A. actinomycetemcomitans ATCC 29522 and P. gingivalis ATCC 33277 were tested with the species-specific primers. Ubiquitous primers were also used as a positive control for the PCR amplification. For each set of primers, PCR was performed on sterile water to check for DNA contamination (negative controls). The specificities of all primers were previously investigated11-14 and primer sequences were compared with similar sequences of the reference organisms by BLAST search (http://ncbi.nlm.nih.gov/blast/). To further test the specificity of the primers, total DNA from both aforementioned bacterial strains was used with each of the primer pairs12,15.
The sequences of primers for P. gingivalis were designed to amplify a 404 base pair (sense primer (5'-3'): AGGCAGCTTGCCATACTGCG, antisense primer (5'-3'): ACTGTTAGCAACTACCGATGT), and for A. actinomycetemcomitans a 557 base pair (sense primer (5'-3'): ATGCCAACTTGACGTTAAAT, antisense primer (5'-3'): AAACCCATCTCTGAGTTCTTCTTC). Ubiquitous primers were used as a positive control for bacterial presence through PCR reaction and were designed to amplify a 602 base pair (sense primer (5'-3'): GATTAGATACCCTGCTAGTCCAC, antisense primer (5'-3'): CCCGGGAACGTATTCACCG). The primer sequences were described by Ashimoto et al.12.
PCR amplification products (9 µL) were analyzed by 2% agarose gel electrophoresis. The agarose gels were stained with 0.5 µg/ml ethidium bromide and photographed under ultraviolet light. A 100 bp DNA ladder served as the molecular weight marker.
The detection limit of the PCR method was determined using DNA from known numbers of target organisms (10-107 cells per PCR as determined by viable cell counts).
The Institutional Experimentation Committee for Human Experiments approved the protocol for this study (protocol # 019/2006).
RESULTS
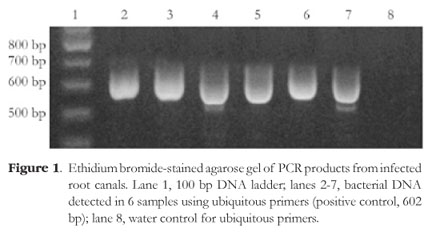
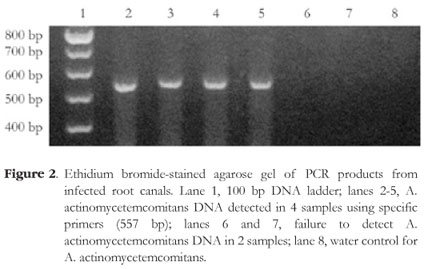
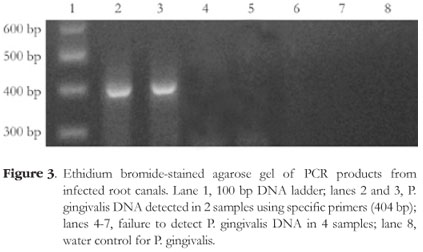
All tested samples contained the amplicon of the ubiquitous bacterial primers (Figure 1). A. actinomycetemcomitans was detected in 16.6% of the cases (4 of 24) and P. gingivalis was detected in 8.3% of the cases (2 of 24). Figures 2 and 3 depict representative amplicons from PCR analysis of the presence of A. actinomycetemcomitans and P. gingivalis, respectively. Reference DNA and clinical samples that were positive for A. actinomycetemcomitans and P. gingivalis showed only one band of 557 bp and 404 bp, respectively.
DISCUSSION
Oral mucous membranes and teeth are colonized by a variety of microorganisms, mostly bacteria, but the vital dental pulp is normally sterile. The most common initiators of pulpal destruction are bacteria. Studies have examined the use of PCR to identify endodontic pathogens in root canals5,9,11,16.
PCR technique has enabled the detection of bacterial species that are difficult or even impossible to culture1-4. In addition, PCR is faster, much more sensitive, and more accurate than culture4,10-11. Its use in endodontics to investigate the microbiota of root canals has expanded the knowledge on the bacteria involved in the pathogenesis of periradicular diseases1,9,11,17-18.
Cultures require at least an 8 h incubation of the sample in the culture medium followed by biochemical and other tests to identify the microorganism. The time required for identification can be even longer for slow-growing microorganisms or samples with low microbial counts. When traditional culture methods are used, laboratories may need 7-14 days to identify anaerobic bacteria, while PCR can provide information in only a few days4.
Other studies have also found a similar prevalence of the pathogens A. actinomycetemcomitans and P. gingivalis in root canals of patients without cleft lip and palate1,3,6,9,11,17-18. These pathogens, A. actinomycetemcomitans and P. gingivalis, are found in endodontic infections more often when the infections are investigated with PCR than with cultures1,3-7,9,11,17-18. This study has demonstrated that PCR is an efficient method for detecting the presence of these bacteria in infected root canals of cleft lip and palate patients.
CONCLUSION
Molecular biology techniques may provide insights into the complete and complex microflora of the oral cavity in general, and particularly of infected root canals. PCR has contributed significantly to the knowledge of the root canal microbiota by allowing the identification of endodontic pathogens. This may also contribute to the development of improved treatment strategies for cleft lip and palate patients.
Collaborators
TCS PEREIRA was responsible for the laboratory work and writing of the article. TM OLIVEIRA helped in the laboratory, in collecting samples and writing the article. VT SAKAI and TJ DIONÍSIO helped in the laboratory. RP HUSSNE helped in the collection of samples. CK NISHIYAMA provided all the clinical support. MAAM MACHADO provided the material used in the study. CF Santos was the supervisor of the study at USP School of Dentistry and wrote the article in English.
REFERENCES
1. Siqueira JF Jr, Rôças IN, Oliveira JCM, Santos KRN. Detection of putative oral pathogens in acute periradicular abscesses by 16S rDNA-directed polymerase chain reaction. J Endod. 2001;27(3):164-7. [ Links ]
2. Conrads G, Gharbia SE, Gulabivala K, Lampert F, Shah HN. The use of a 16rDNA directed PCR for the detection of endodontopathogenic bacteria. J Endod. 1997;23(7):433-8. [ Links ]
3. Rolph HJ, Lennon A, Riggio MP, Saunders WP, MacKenzie D, Coldero L, et al. Molecular identification of microorganisms from endodontic infections. J Microb. 2001;39(9):3282-9. [ Links ]
4. Siqueira JF Jr, Rôças IN. PCR methodology as a valuable tool for indentification of endodontic pathogens. J Dent. 2003;1(5):333-9. [ Links ]
5. Siqueira JF Jr, Rôças IN. Molecular detection and identification of Synergistes phylotypes in primary endodontic infections. Oral Dis. 2007;13(4):398-401. [ Links ]
6. Siqueira JF Jr, Rôças IN. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004,97(1):85-94. [ Links ]
7. Siqueira JF Jr, Rôças IN, Moraes SR, Santos KR. Direct amplification of rRNA gene sequences for identification of selected oral pathogens in root canal infections. Int Endod J. 2002;35(4):345-51. [ Links ]
8. Pitt TL, Saunders NA. Molecular bacteriology: a diagnostic tool for the millennium. J Clinical Pathol. 2000;53(1):71-5. [ Links ]
9. Siqueira JF Jr, Rôças IN, Cunha CD, Rosado AS. Novel bacterial phylotypes in endodontic infections. J Dent Res. 2005;84(6):565-9 [ Links ]
10. Santos CF, Sakai VT, Machado MAAM, Schippers DN, Greene AS. Reverse transcription and polymerase chain reaction: principles and applications in dentistry. J Appl Oral Sci. 2004;12(1):1-11. [ Links ]
11. Oliveira JCM, Siqueira JF Jr, Alves GB, Hirata Jr R, Andrade AFB. Detection of porphyromonas endodontalis in infected root canals by 16S rRNA gene-directed polymerase chain reaction. J Endod. 2000;26(12):729-32. [ Links ]
12. Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11(4):266-73. [ Links ]
13. Kimura S, Ooshima T, Takiguchi M, Sasaki Y, Amano A, Morisaki I, et al. Periodontopathic bacterial infection in childhood. J Periodontol. 2002;73(1):20-6. [ Links ]
14. Ooshima T, Nishiyama N, Hou B, Tamura K, Amano A, Kusumoto A, et al. Occurrence of periodontal bacteria in healthy children: a 2-year longitunial study. Community Dent Oral Epidemiol. 2003;31(6):417-25. [ Links ]
15. Umeda M, Contreras A, Chen C, Bakker I. The utility of whole saliva to detect the oral presence of periodontopathic bacteria. J Periodontol. 1998;69(7):828-33. [ Links ]
16. Nandakumar R, Whiting J, Fouad AF. Identification of selected respiratory pathogens in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;106(1):969-75. [ Links ]
17. Bate AL, Ma JKC, Ford TRP. Detection of bacterial virulence genes associated with infective endocarditis in infected root canals. Int Endod J. 2000;33(3):194-203. [ Links ]
18. Bogen G, Slots J. Black-pigmented anaerobic rods in closed periapical lesions. Int Endod J. 1999,32(3):204-10. [ Links ]
Recebido em: 5/7/2008
Versão final reapresentada em: 23/10/2008
Aprovado em: 1/4/2009
* Correspondência para / Correspondence to: TM OLIVEIRA. E-mail: <marchini@usp.br>